Hands on spectral clustering in R
Spectral clustering is a class of techniques that perform
cluster division using eigenvectors of the similarity matrix. The division is
such that points in the same cluster should be highly similar and points in
different clusters should have highly dissimilar. Thus, spectral clustering is a
non-parametric method of clustering. One of the advantages of this clustering
mechanism is that it is not affected by outliers or noise and performs fast!
Let’s open R
We
would look at three packages in R but first, let’s setup the system
- Go to https://www.r-project.org and download R for your system
- After installing R, install R-studio from https://www.rstudio.com/
- After installing, Open R studio and install the spectral clustering packages in a new R script by executing the lines:
These lines install the three packages SamSPECTRAL, kknn and kernlab in R
SamSPECTRAL
It contains a function by the same name (SamSPECTRAL) which
performs spectral clustering. Let’s try out some code first.
The performance of the package depends upon some assumptions
– values of separation factor, dimension and normal.sigma. The dimension vector
should be the set of dimensions used for clustering (used all 3 dimensions
here). The sigma value determines how many spectral clusters should be detected
whereas the separation factor is used for determining minimum distance between
clusters. The better the estimates of these variables, the better the
clustering. Here separation factor of about 0.7 is more optimal than the
example used. Similarly, sigma value of 250 gives better results and helps
reduce the neutral class. Another importance of this package is its performance for large datasets. Had the same dataset used for spectral clustering in any other package, it would take a lot of time to complete.
Kernlab
Kernlab is a kernel package and
includes many functions, one of which is spectral clustering. It also contains
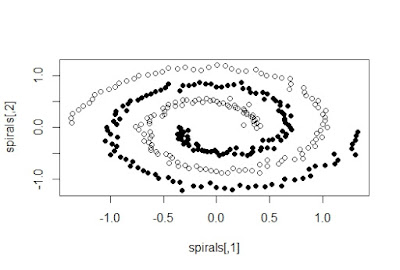
datasets of which we would use the spirals data set. It is a two-dimensional
data set with 300 data points. When plotted, it appears as two spirals with
Gaussian noise in each data point. We have a function specc which runs spectral
clustering. Let’s have a look at code:
Though not used in the
example. Kernlab specc function can handle null values quite well. At the same
time, it can apply many kernel functions. Hence we have a greater flexibility
over the clustering hence the accurate results even in a spiral type data
arrangement as this. However, as the method is so complex, its performance
decreases as the data size increases. These would be the cases when SamSPECTRAL
would outsmart specc model. Do look at the help documentation of each package
to know more about the functioning.
Kknn
The final package this
blog article covers is kknn package. Unlike the previous two, this package uses
k-nearest neighbor technique to generate the similarity matrix rather than
k-means. This time we have the function specClust. The last piece of code looks
like this:
The iris dataset already
has 3 classes of 50 members each and could help us to check the accuracy of
clustering:
This matrix shows that
we have correctly classified all ‘setosa’ type points but made errors of 2 and
4 in ‘versicolor’ and ‘virginica’ respectively. Hence the total errors are
6/150 (4%)
Comparison
We first saw kknn
package which uses an entirely different approach of k-nn to cluster datasets.
On the other hand, SamSPECTRAL is more suited to larger datasets than kernlab.
Each of the packages is thus suitable for different types of datasets. Had the
iris dataset used over kernlab, it would have produced a much poor clustering
than kknn did. Similarly, the dataset used for SamSPECTRAL would cause
performance issues on the other two. Moreover, Spectral clustering can be
implemented in many more ways than these packages alone. Hence, we should always have a first look at
the data and use the suitable method accordingly.
Fellow coders, feel free
to comment in case of any doubts. You can also contact me offline.
References:
- Codes are available at : google drive
- SamSPECTRAL: A Modied Spectral Clustering Method for Clustering Flow Cytometry Data [pdf]
- kernlab – An S4 Package for Kernel Methods in R [pdf]
- specClust {kknn} http://www.inside-r.org/packages/cran/kknn/docs/specClust






i am getting error so kindly advice to solve it
ReplyDeleten .Call("R_igraph_arpack", func, extra, options, env, sym, PACKAGE = "igraph") :
At arpack.c:944 : ARPACK error, Maximum number of iterations reached
In addition: Warning message:
In .Call("R_igraph_arpack", func, extra, options, env, sym, PACKAGE = "igraph") :
At arpack.c:776 :ARPACK solver failed to converge (1001 iterations, 0/7 eigenvectors converged